Questions and answers for dimensionality reductions
1. What is dimensionality reduction?
When we have a dataset with multiple input features, we know the model will overfit. To reduce input feature space, we can either drop or extract features, this is basically a dimension reduction.
Now let’s discuss more about both techniques.
- Drop irrelevant, redundant features as they do not contribute to the accuracy of the predictive problem. When we drop such input variable, we lose information stored in these variables.
- We can create a new independent variable from existing input variables. This way we do not lose the information in the variables. This is feature extraction
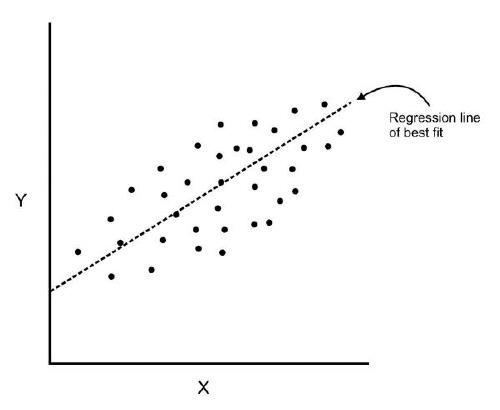
2. Explain Principal Component Analysis?
When we have a large dataset of correlated input variables and we want to reduce the number of input variables to a smaller feature space. while doing this we still want to maintain the critical information. We can solve this by using Principal Component Analysis-PCA.
Now let’s understand the PCA features in little bit more details.
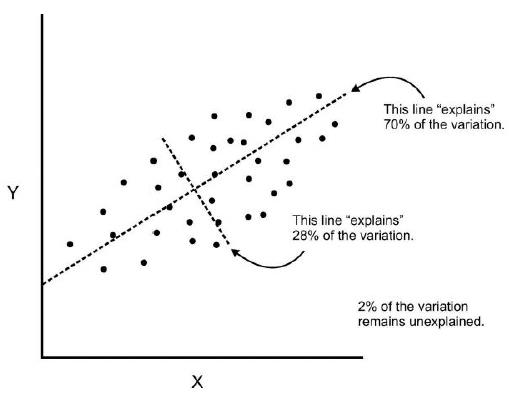
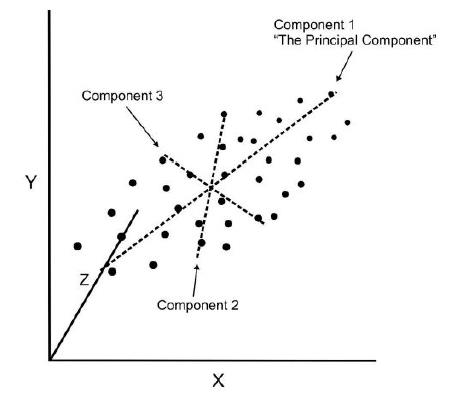
PCA reduce dimensionality of the data using feature extraction. It does this by using variables that help explain most variability of the data in the dataset.
PCA removes redundant information by removing correlated features. PCA creates new independent variables that are independent from each other. This takes care of multicollinearity issue.
PCA is an unsupervised technique. It only looks at the input features and does not take into account the output or the target variable.
3. Importance and limitation of Principal Component Analysis?
Following are the advantages of PCA
- Removes Correlated Features – To visualize our all features in data, we must reduce the same in data, to do that we need to find out the correlation among the features (correlated variables). Finding correlation manually in thousands of features is nearly impossible, frustrating and time-consuming. PCA does this for you efficiently.
- Improve algorithm performance – With so many features, the performance of your algorithm will drastically degrade. PCA is a very common way to speed up your Machine Learning algorithm by getting rid of correlated variables which don’t contribute in any decision making.
- Improve Visualization – It is very hard to visualize and understand the data in high dimensions. PCA transforms a high dimensional data to low dimensional data (2 dimension) so that it can be visualized easily.
Following are the limitation of PCA
- Independent variable become less interpretable – After implementing PCA on the dataset, your original features will turn into Principal Components. Principal Components are the linear combination of your original features. Principal Components are not as readable and interpretable as original features.
- Data standardization is must before PCA – You must standardize your data before implementing PCA, otherwise PCA will not be able to find the optimal Principal Components.
- Information loss – Although Principal Components try to cover maximum variance among the features in a dataset, if we don’t select the number of Principal Components with care, it may miss some information as compared to the original list of features.
4. What is t-SNE and How to apply t-SNE ?
t-Distributed Stochastic Neighbor Embedding (t-SNE) is an unsupervised, non-linear technique primarily used for data exploration and visualizing high-dimensional data. In simpler terms, t-SNE gives you a feel or intuition of how the data is arranged in a high-dimensional space.
Let’s understand each and every term in details.
Scholastic – Not definite but random probability
Neighbourhood – Concerned only about retaining the structure of neighbourhood points.
Embedding – It means picking up a point from high dimensional space and placing it into lower dimension
5. How to apply t-SNE ?
Basically, it measure similarities between points in the high dimensional space.
Let’s see below image and try to understand the algorithm.
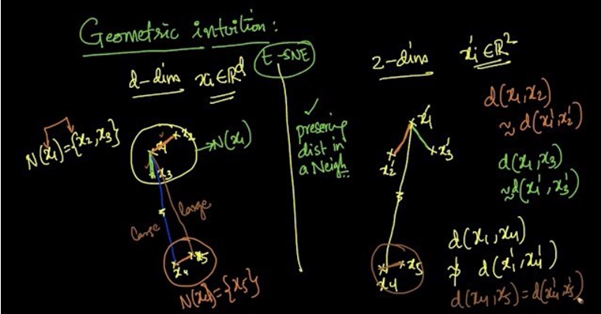
Suppose we are reducing d-dimensional data into 2-dimensional data using t-SNE.
From the above picture we can see that x2 and x3 are in the neighborhood of x1 [N(x1) = {x2, x3}] and x5 is in the neighborhood of x4 [N(x4) = {x5}].
As t-SNE preserves the distances in a neighborhood,
d(x1, x2) ≈ d(x’1, x’2)
d(x1, x3) ≈ d(x’1, x’3)
d(x4, x5) ≈ d(x’4, x’5)
For every point, it constructs a notion of which other points are its ‘neighbors,’ trying to make all points have the same number of neighbors. Then it tries to embed them so that those points all have the same number of neighbors.
6. What is Crowding problem?
Sometimes it is impossible to preserve the distances in all the neighbourhoods. This problem is called Crowding Problem or When we model a high-dimensional dataset in 2 (or 3) dimensions, it is difficult to segregate the nearby datapoints from moderately distant datapoints and gaps can not form between natural clusters.
For example, when a data point, ‘x’ is a neighbor to 2 data points that are not neighboring to each other, this may result in losing the neighborhood of ‘x’ with one of the data points as t-SNE is concerned only within the neighborhood zone.
7. How to interpret t-SNE output?
There are 3 parameters
a) Steps: number of iterations.
b) Perplexity: can be thought of as the number of neighboring points.
c) Epsilon: It is for data visualization and determines the speed which it should be changed.